Quantile layer for serial axes coordinate
geom_serialaxes_quantile.RdDraw a quantile layer for serial axes coordinate.
Don't be confused with geom_quantile() which is a quantile regression. See examples.
geom_serialaxes_quantile(
mapping = NULL,
data = NULL,
stat = "serialaxes",
position = "identity",
...,
quantiles = seq(0, 1, 0.25),
axes.sequence = character(0L),
merge = TRUE,
na.rm = FALSE,
orientation = NA,
show.legend = NA,
inherit.aes = TRUE
)
stat_serialaxes_quantile(
mapping = NULL,
data = NULL,
geom = "serialaxes_quantile",
position = "identity",
...,
axes.sequence = character(0L),
merge = TRUE,
quantiles = seq(0, 1, 0.25),
scaling = c("data", "variable", "observation", "none"),
axes.position = NULL,
na.rm = FALSE,
orientation = NA,
show.legend = NA,
inherit.aes = TRUE
)Arguments
- mapping
Set of aesthetic mappings created by
aes(). If specified andinherit.aes = TRUE(the default), it is combined with the default mapping at the top level of the plot. You must supplymappingif there is no plot mapping.- data
The data to be displayed in this layer. There are three options:
If
NULL, the default, the data is inherited from the plot data as specified in the call toggplot().A
data.frame, or other object, will override the plot data. All objects will be fortified to produce a data frame. Seefortify()for which variables will be created.A
functionwill be called with a single argument, the plot data. The return value must be adata.frame, and will be used as the layer data. Afunctioncan be created from aformula(e.g.~ head(.x, 10)).- stat
The statistical transformation to use on the data for this layer, either as a
ggprotoGeomsubclass or as a string naming the stat stripped of thestat_prefix (e.g."count"rather than"stat_count")- position
Position adjustment, either as a string naming the adjustment (e.g.
"jitter"to useposition_jitter), or the result of a call to a position adjustment function. Use the latter if you need to change the settings of the adjustment.- ...
Other arguments passed on to
layer(). These are often aesthetics, used to set an aesthetic to a fixed value, likecolour = "red"orsize = 3. They may also be parameters to the paired geom/stat.- quantiles
numeric vector of probabilities with values in [0,1]. (Values up to 2e-14 outside that range are accepted and moved to the nearby endpoint.)
- axes.sequence
A vector to define the axes sequence. In serial axes coordinate, the sequence can be either determined in
mapping(functionaes()) or byaxes.sequence. The only difference is that themappingaesthetics will omit the duplicated axes (check examples ingeom_serialaxes).- merge
Should
axes.sequencebe merged with mapping aesthetics as a single mappingunevalobject?- na.rm
If
FALSE, the default, missing values are removed with a warning. IfTRUE, missing values are silently removed.- orientation
The orientation of the layer. The default (
NA) automatically determines the orientation from the aesthetic mapping. In the rare event that this fails it can be given explicitly by settingorientationto either"x"or"y". See the Orientation section for more detail.- show.legend
logical. Should this layer be included in the legends?
NA, the default, includes if any aesthetics are mapped.FALSEnever includes, andTRUEalways includes. It can also be a named logical vector to finely select the aesthetics to display.- inherit.aes
If
FALSE, overrides the default aesthetics, rather than combining with them. This is most useful for helper functions that define both data and aesthetics and shouldn't inherit behaviour from the default plot specification, e.g.borders().- geom
The geometric object to use to display the data, either as a
ggprotoGeomsubclass or as a string naming the geom stripped of thegeom_prefix (e.g."point"rather than"geom_point")- scaling
one of
data,variable,observationornone(not suggested the layout is the same withdata) to specify how the data is scaled.- axes.position
A numerical vector to determine the axes sequence position; the length should be the same with the length of
axes.sequence(or mappingaesthetics, see examples).
Examples
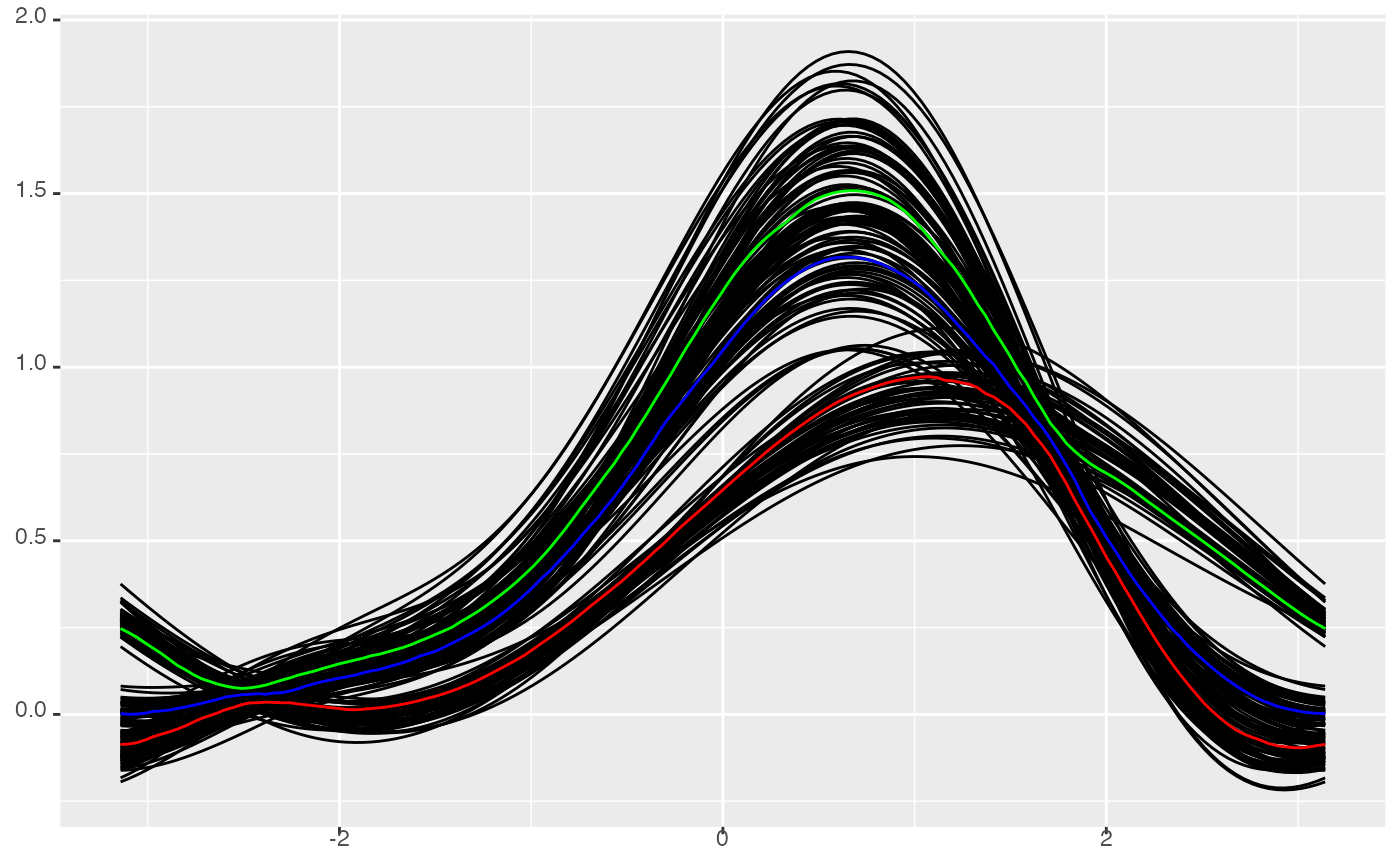
# lower quantile, median and upper quantile
p <- ggplot(iris, mapping = aes(Sepal.Length = Sepal.Length,
Sepal.Width = Sepal.Width,
Petal.Length = Petal.Length,
Petal.Width = Petal.Width)) +
geom_serialaxes(stat = "dotProduct") +
geom_serialaxes_quantile(stat = "dotProduct",
quantiles = c(0.25, 0.5, 0.75),
colour = c("red", "blue", "green"))
p