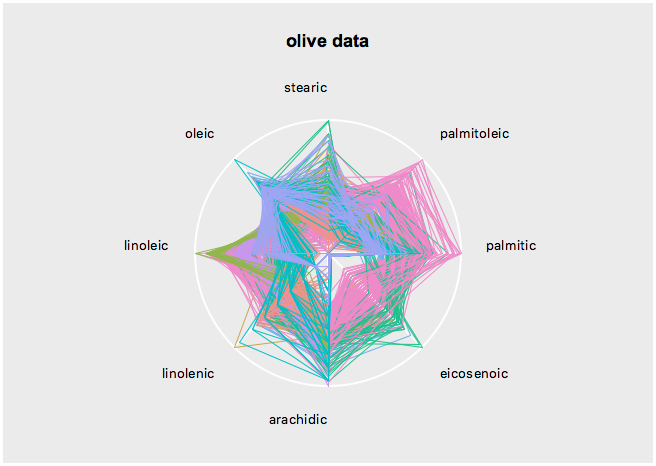

s <- l_serialaxes(data=oliveAcids, color=olive$Area, title="olive data")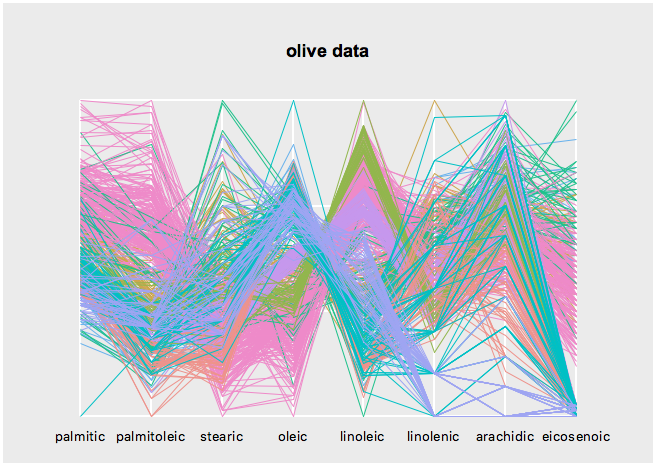
s['axesLayout'] <- 'parallel'Get the state names with
states <- l_info_states(s)
names(states)Query a state, say sequence, as follows
s['sequence']Change a state, say again sequence, as follows
s['sequence'] <- names(olive)[c(2:10)]alternatively, and more efficient if you modify more than one state, use
l_configure(s, sequence=c('stearic', 'linoleic', 'palmitic', 'arachidic'))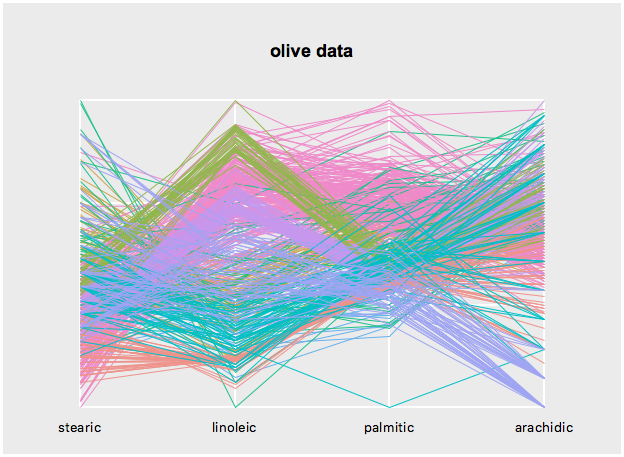
When creating a plot you may specify any state at plot creation
s1 <- l_serialaxes(data=olive, color=olive$Area, title="olive data",
sequence=names(olive)[2:10])details on a state, say sequence, is easily had with
states <- l_info_states(s)
states$sequenceand a particular field
states$sequence$descriptionThe scaling state defines how the data is scaled. The axes display 0 at one end and 1 at the other. For the following explanation assume that the data is in a nxp dimensional matrix. The scaling options are then