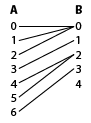
many to one is combined by a logical OR
All of loon's plots can be linked so that some of their n-dimensional states are synchronized. For a linked scatterplot, the states that are synchronized by default (i.e. linked states) are the color, size, selected and active states. Two or more plots are linked if they share the same string in their linkingGroup state.
loon's standard linking model is fairly flexible and the user can choose
n dimensional states should be synchronizedn dimensional states from one display to another display.We allow only one-to-one synchronization. For example, selecting a point and have it's k nearest neighbors automatically selected in the same display is not possible. Also, it is not possible with the standard linking mechanism to have one selected point in display A result in selecting multiple points in display B. For cases where the standard model is not sufficient it is possible to implement custom linking rules with state bindings.
The standard linking model depends on the two states linkingGroup and linkingKey.
Displays are linked if their linkingGroup state contains the same string.
none is a keyword; if linkingGroup equals none then the plot is not linked.For example, for
p1 <- l_plot(iris[,1:2], linkingGroup='iris')
p2 <- l_plot(iris[,2:3], linkingGroup='iris')
p3 <- l_plot(iris[,3:4], linkingGroup='none')
h <- l_hist(iris[,1], linkingGroup='iris')the plots p1, p2, and h are linked to each other and p3 is not linked to any other display.
The states of a plot that are linked can be queried with
l_getLinkedStates(p1)Usually the linked states are color, selected, active and size, if they exist for the particular plot. Histograms, for example, have no size state. Note that between displays only states with the same state name can be linked. To set the (n-dimensional!) linked states use
l_setLinkedStates(p1, c('color', 'active'))If display A is linked with display B then
A and B share the same linking group (i.e. string in the linkingGroup state)A and B are synchronizedi'th elements of the states in A are synchronized with the i'th elements of the same states in display B for all i in the intersection of the point indices of display A and BHence, for the above example where n=150 (i.e. the number of data points) when selecting the i'th point in p1 then loon will set the selected state for the i'th point in p2 and h to TRUE, for any i in {1,...,150}. If a further display gets created with linkingGroup iris but a different value for n, say n=4, then only the first for elements of that plot get synchronized between that display and p1, p2, and h. For example, for
p4 <- l_plot(1:4,1:4, linkingGroup='iris')any change in p4 for any of the linked states will only affect the first 4 elements in p1, p2 and h. Or vice versa, only the states of the first for elements in p1, p2 and h will ever have an effect on p4.
It is possible to change the mapping of which elements should be synchronized between displays. The n dimensional linkingKey state controls which elements get synchronized between the linked displays. That is for two linked displays A and B
element i for the linked states in display A is kept in sync with element j for the linked states in display B if there is an i and j for which the i'th element in the linkingKey state of display A contains exactly the same string as the element j in the linkingKey state of display B
the linkingKey state for a display must contain unique elements
By default, the linkingKey contains an n dimensional vector with 0,1,...,n-1.
Both, the linkingGroup state and the linkingKey state can be changed at run time. Changing the p2 in the above example to have linkingGroup contain the iris2 string, you can run
l_configure(p2, linkingGroup='iris2')now only p1 and h are linked.
If a plot's, say display A, linkingGroup is changed to a linking group that already has linked members, say display B, C, and D, then one must specify whether the initial synchronization of the linked states should be a push, i.e. A > B, C, D, or a pull, i.e. A < B, C, D. This is done with the sync argument:
l_configure(p3, linkingGroup='iris2', sync='pull')The sync argument must also be used if the linkingKey state gets changed of a plot that has linked displays. For example, to link the points in p4 with the last 4 points in the iris data use
l_configure(p4, linkingKey=c(146,147,148,149), sync='push')Note that using the default linkingKey results in the fastest linking.
If more flexible linking rules are needed, one can implement state bindings.
For example, for the following displays pa and pb
pa <- l_plot(x=1:7, y=1:7, title="A")
pb <- l_plot(x=1:5, y=1:5, title="B")Assume the linking:

many to one is combined by a logical OR
pa2pb <- function() {
sa <- pa['selected']
sb <- pb['selected']
pb['selected'] <- c(any(sa[1:3]), sa[4], any(sa[5:6]), sa[7], sb[5])
}
pb2pa <- function() {
sb <- pb['selected']
pa['selected'] <- c(rep(sb[1],3), sb[2], rep(sb[3],2), sb[4])
}
l_bind_state(pa, 'selected', pa2pb)
l_bind_state(pb, 'selected', pb2pa)This will not end in an endless loop of evaluating state bindings as after the sequence pa2pb - pb2pa - pa2pb or the pb2pa - pa2pb - pb2pa the system will be in equilibrium with respect to these functions. Remember that state bindings do not get evaluated if a configure call does not effectively change the state. If, however, the custom linking does not result in an equilibrium after the first change, then you may need to add a further variable, say busy, to avoid multiple iterations -- or an infinite loop--. Assume the linking
directions are indicated by arrows
pa2 <- l_plot(x=1:7, y=1:7, title="A 2")
pb2 <- l_plot(x=1:5, y=1:5, title="B 2")
busy <- FALSE
a2b <- function() {
if(!busy) {
busy <<- TRUE
sa <- pa2['selected']
l_configure(pb2, selected=!sa[1], which_n=1)
busy <<- FALSE
}
}
b2a <- function() {
if(!busy) {
busy <<- TRUE
sb <- pb2['selected']
l_configure(pa2, selected=!sb[1], which_n=1)
busy <<- FALSE
}
}
l_bind_state(pa2, 'selected', a2b)
l_bind_state(pb2, 'selected', b2a)which_n will not be changed in the configure call.Without the variable busy this would end in an infinite loop one the selected state gets changed of either display.
Note that in the previous two examples the linking functions scope for the plot handles. To avoid unintended side effects, for example, by overwriting or deleting a plot handle it is advisable to wrap the linking functions in another R function.
For example, for the first example the following setup would be better
myLinking <- function(pa, pb) {
pa2pb <- function() {
sa <- pa['selected']
sb <- pb['selected']
pb['selected'] <- c(any(sa[1:3]), sa[4], any(sa[5:6]), sa[7], sb[5])
}
pb2pa <- function() {
sb <- pb['selected']
pa['selected'] <- c(rep(sb[1],3), sb[2], rep(sb[3],2), sb[4])
}
c(l_bind_state(pa, 'selected', pa2pb),
l_bind_state(pb, 'selected', pb2pa))
}Now any two plots can be linked with the particular linking rule defined in myLinking as follows
plotA <- l_plot(x=1:7, y=1:7, title="plot A")
plotB <- l_plot(x=1:5, y=1:5, title="plot B")
myLinking(plotA, plotB)Note that the myLinking function call environment does not get garbage collected as the bindings refer to the two functions pa2pb and pb2pa. You can read more about environments in Hadley's Advanced R book.